Protocol for assembling annealed DNA oligonucleotides and a double-stranded DNA vector using NEBuilder HiFi DNA Assembly (NEB #E2621)
The following method allows you to anneal short overlapping DNA oligos (generally 60 nucleotides (nt) each) to assemble a longer double-stranded DNA (dsDNA) fragment. The annealed nicked dsDNA fragment can then be combined and assembled with a linearized vector fragment. This annealed oligo protocol provides an alternative to short, synthesized dsDNA, such as gBlocks™.
This protocol is recommended for the assembly of the following types of DNA fragments:
- Annealed short DNA oligos forming a nicked dsDNA fragment
- dsDNA vector linearized by PCR or restriction digest
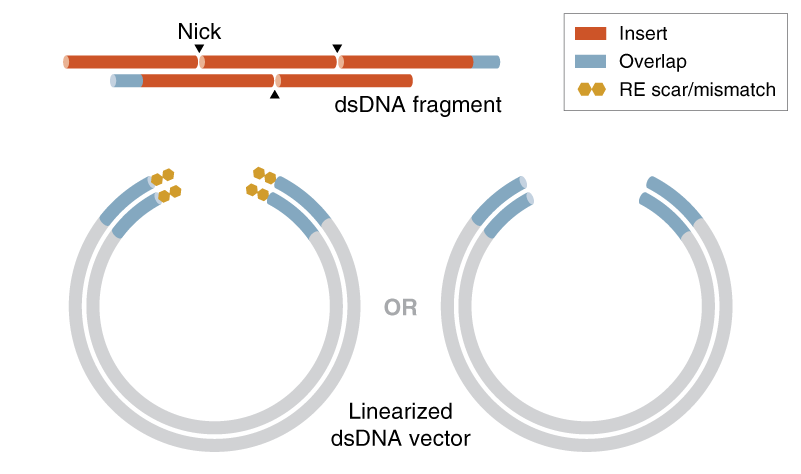
Annealed DNA Oligo Design
Short, annealed ssDNA oligos (60 nt each) should be designed with 30 nt overlaps with adjacent complementary oligos. When annealed, the overlapping oligos will form a nicked dsDNA fragment with no gaps, and ssDNA vector overlaps at each end. Please note that DNA oligos with 5’ phosphates are not required.
DNA Quantities
This protocol uses a 1:50 (vector:insert) molar ratio with 0.02 picomoles of vector and 1 picomole of annealed oligos.
Protocol
- Prepare oligos for annealing by adding 1 ul of each oligo (100 μM stock) to a final concentration of 0.2 μM (0.2 pmol/μl) using 1X NEBuffer r2.1*. This can be done by combining 1 µl of each 100 μM oligo stock in a single tube with an appropriate volume of buffer for a total volume of 500 μl.
- Heat the oligo mixture solution at 100°C for 3 min and allow to cool at room temperature for 20 min. The annealed oligos are ready to assemble.
- Set-up the following reaction on ice:
- Incubate the reaction at 50°C in a thermocycler for 60 min. Transform 2 μl of assembled mix into 50 μl of NEB 5-alpha Competent E. coli (High Efficiency) (NEB #C2987) following the recommended protocol.
| Component | 500 µl Volume | Final Conc. Or Amount |
|---|---|---|
| Overlapping Oligos (100 μM stock concentration) | 1 μl of each oligo, for a total of X μl | 0.2 μM (0.2 pmol/μl) of each oligo |
| 1X NEBuffer r2.1* | Buffer volume = (500 μl - X μl) | 1X |
*Note: you can also use TE buffer (10 mM Tris, 0.1 mM EDTA; pH 8.0) supplemented with 50 mM NaCl as an annealing buffer
| Component | 20 µl Reaction | Final Conc. Or Amount |
|---|---|---|
| Annealed Oligo Mixture (0.2 pmol/μl) |
5 μl | 1 pmol (1:50 vector:insert ratio) |
| Linearized Vector (0.02 pmol/μl)** | 1 μl | 0.02 pmol |
| Nuclease-free Water | 4 μl | |
| NEBuilder HiFi DNA Assembly Master Mix 2X | 10 μl | 1X |
** NEBioCalculator (NEBioCalculator.neb.com) can help with DNA mass to molar quantity conversions for both ssDNA and dsDNA.
Note: If you are working with large plasmids >10 kb in size we recommend NEB® 10-beta Competent E. coli (High Efficiency) (NEB #C3019H). If your plasmid or insert contain repetitive sequences, we recommend NEB® Stable Competent E. coli (High Efficiency) (NEB #C3040H).
